- Home
- Introduction
- Downloads
- Example
-
User Guide
Index
 System requirement
System requirement  Installation
Installation  Memory configuration, reducing memory usage
Memory configuration, reducing memory usage  Updating annotation databases
Updating annotation databases  Main user interface
Main user interface  Data inputs
Data inputs  Creating a project
Creating a project  Annotating a project
Annotating a project  Using user annotation track [GFF3/BED]
Using user annotation track [GFF3/BED]  Analyzing a project
Analyzing a project  Selecting genes or regions
Selecting genes or regions  Exome or targeted capture sequencing A command line tool Version history
Exome or targeted capture sequencing A command line tool Version history  Updating to the latest version FAQ Requests & discussions License
Updating to the latest version FAQ Requests & discussions License
- Screenshot
- Java Dev
- Plug-ins
- Visitors
Using this software
0. For impatient users1. Data inputs
2. Create a project
3. Annotate a project
4. Filter for quality scores
5. Main user interface
6. SVA genome browser
7. SVA tables
8. Selecting genes or regions
9. Analysis
10. Exome or targeted capture sequencing
A command line tool
FAQ
Requests and discussions

Create a new project
SVA manages its projects through a project file. This project file is in a flat text format, and must have a file name extension .gsap .
There are two ways to create a new SVA project: (1) through menu "File -> Create a new SVA project"; or (2) make a copy of an existing project (for example, the example project included in the SVA package), and modify the input and output files. Sometimes we found that options 2 is more productive, particularly for differernt projects with same participants and same data files.
I will describe option 1 in this page.
Step 1. Create a new .gsap project file
Click on menu "File -> Create a new SVA project", and you will be asked where to save the new project:
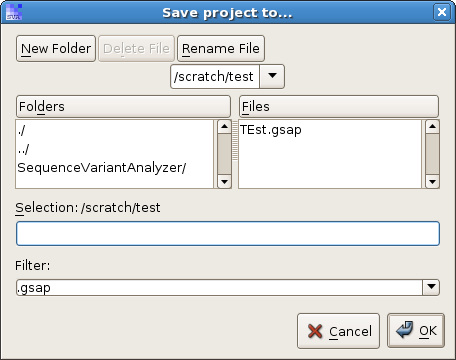
Specify a new project file, and a new project builder will pop up:
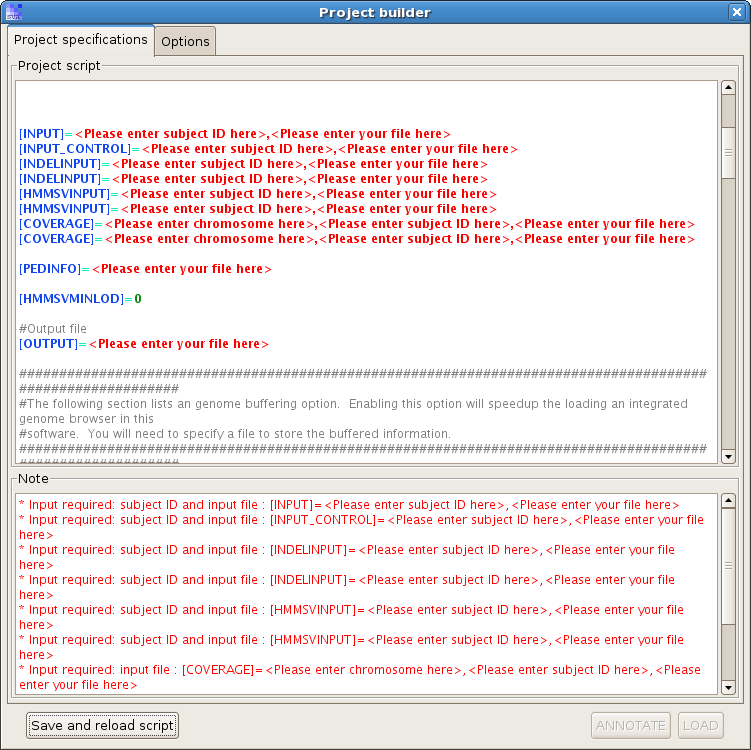
Don't be concerned about all the red flags in the project builder - all it wants is the data inputs you already generated. But do note that if you see anything red in the "Project script" or "Note" area, the project annotation cannot proceed. The red flags indicate that something is missing, or the specified data files do not exist, etc.
You need to type in the subject IDs, location of files, etc., as required by the project builder. Please be cautious with the "[OUTPUT]=" flag. SVA will output to the file you specify in this field. If you specify an existing file, by default SVA will try to load that file directly, instead of overwriting it with a new annotation job. So if you do want a brand new project, delete the old .bin output, or specify a new file to save. You can change this setting by set a switch "[SKIPANNOTATION]=" to "off". In that case SVA will perform a new annotation job no matter the output file exists or not.
Step 2. Performing function annotation
After you type in all the data files, click on button "Save and reload script". Now the new project is saved and you should be able to click on button "ANNOTATE" to perform the main function annotation.
© 2011