- Home
- Introduction
- Downloads
- Example
-
User Guide
Index
 System requirement
System requirement  Installation
Installation  Memory configuration, reducing memory usage
Memory configuration, reducing memory usage  Updating annotation databases
Updating annotation databases  Main user interface
Main user interface  Data inputs
Data inputs  Creating a project
Creating a project  Annotating a project
Annotating a project  Using user annotation track [GFF3/BED]
Using user annotation track [GFF3/BED]  Analyzing a project
Analyzing a project  Selecting genes or regions
Selecting genes or regions  Exome or targeted capture sequencing A command line tool Version history
Exome or targeted capture sequencing A command line tool Version history  Updating to the latest version FAQ Requests & discussions License
Updating to the latest version FAQ Requests & discussions License
- Screenshot
- Java Dev
- Plug-ins
- Visitors
Using this software
0. For impatient users1. Data inputs
2. Create a project
3. Annotate a project
4. Filter for quality scores
5. Main user interface
6. SVA genome browser
7. SVA tables
8. Selecting genes or regions
9. Analysis
10. Exome or targeted capture sequencing
A command line tool
FAQ
Requests and discussions

An introduction to the SVA genome browser displayed sections
There is a general rule for coloring the genome browser displayed sections: if the background of a section is black, it means that the data for this section are from your project. Otherwise the data are from public databases.
There is a list of descriptions on the displayed sections at the end of this page.
You can interact (by mousing clicking or keyboard) with most of the items displayed in the SVA genome browser to obtain detailed information or to navigate to public databases. For details see here >>>
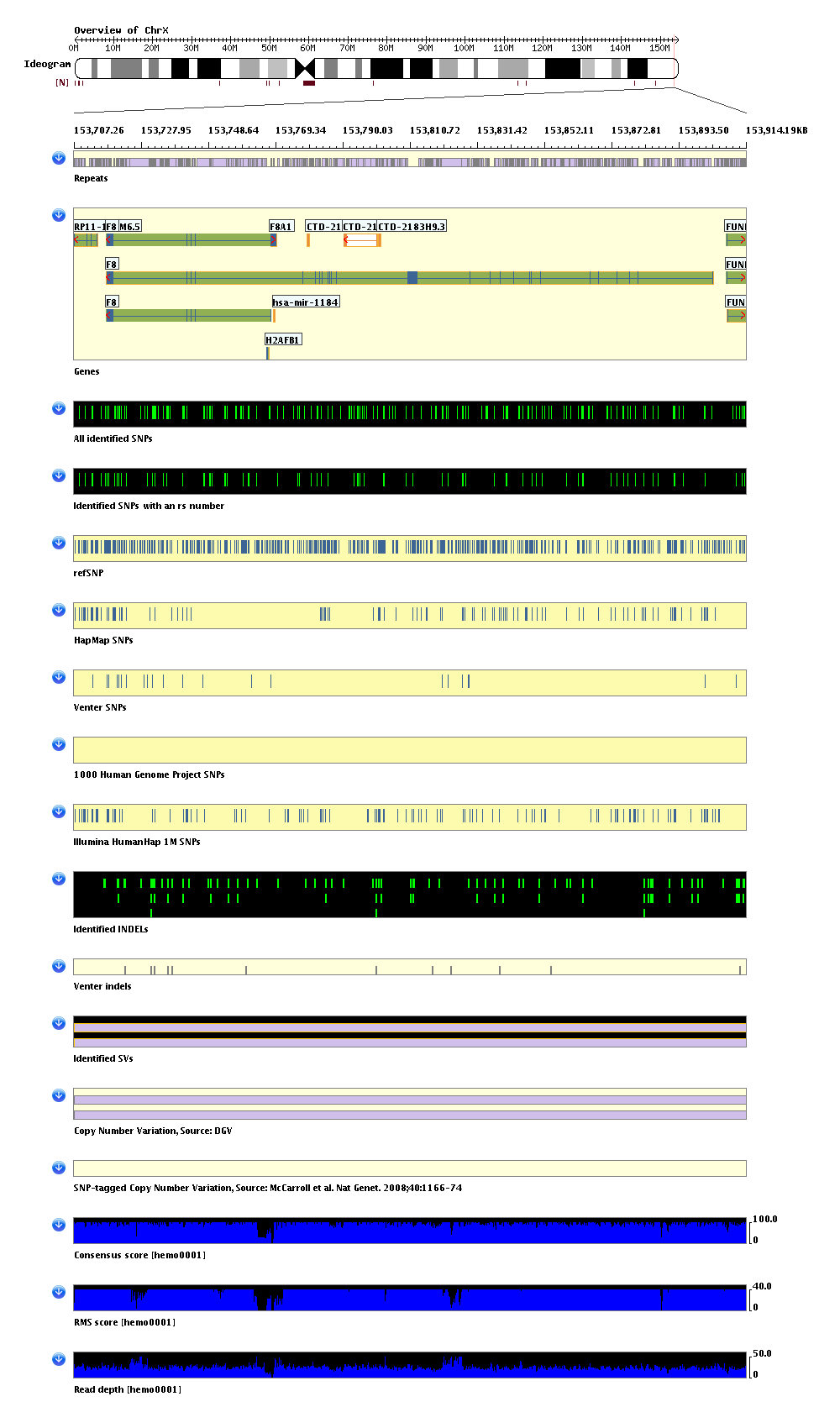 For the figure above, these are the sections displayed from top to bottom:
For the figure above, these are the sections displayed from top to bottom:
| 1. Ideogram overview of the chromosome. The displayed region is highlighed with a red vertical rectangle, or a red vertical line for a small region. |
| 2. Repeats. From RepeatMasker data for build 36. |
| 3. Genes. From Ensembl. Protein-coding genes are plotted as blue (exons) on green (introns). Non protein-coding genes are plotted as orange (exons) on white (introns). Gene coding directions are plotted in red arrows. The most common, or 'canonical' transctipts are plotted with an orange outline (the largest/middle F8 transcript in this example). You may click on transcript for detailed information and navigating to public database (detailed later in this documentation). |
| 4. Identified SNVs. From your project. SNVs are plotted as green lines in black background. Click on SNVs for detailed information (detailed later in this documentation). |
| 5. Identified SNVs with an rs number. From your project. A subset of section 4. |
| 6. Reference SNP. From Ensembl and dbSNP. SNVs are plotted as blue lines in yellow background. Click on SNVs for detailed information and navigating to public database (detailed later in this documentation). |
| 7. HapMap SNP. From HapMap. SNVs are plotted as blue lines in yellow background. Click on SNVs for detailed information and navigating to public database (detailed later in this documentation). |
| 8. Venter's SNP. From J. Craig Venter Institute. Click on SNVs for detailed information (detailed later in this documentation). |
| 9. 1000 Human Genome Project SNPs. From the 1000 Human Genome Project. Click on SNVs for detailed information and navigating to public database if rs# is available (detailed later in this documentation). |
| 10. Illumina Infinium HD Human1M BeadChip SNPs: SNPs included in Illumina Infinium HD Human1M BeadChip (version 1). |
| 11. Identified INDELs. From your project. INDELs are plotted as green rectangles in black background. Click on INDELs for detailed information (detailed later in this documentation). |
| 12. Venter INDELs: From J. Craig Venter Institute. Click on INDELs for detailed information (detailed later in this documentation). |
| 13. Identified SVs. From your project. SVs are plotted as pink rectangles in black background. Click on SVs for detailed information (detailed later in this documentation). |
| 14. DGV SVs. From database of genomic variants (DGV). Click on SVs for detailed information. |
| 15. SNP tagged CNVs. From McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 2008; 40 (10) : 1166-74. |
| 16. Phred-like consensus score. From your project. |
| 17. RMS score. From your project. |
| 18. Read depth. From your project. |
| Visits: |
© 2011
© 2011